1. We're using machine learning algorithms to design new types of microscopes
Recent papers:
K. Kim et al, "Multi-element microscope optimization by a learned sensing network with composite physical layers," Optics Letters 2020.
Project page with data and source code: https://deepimaging.io/me-microscope/
C. L. Cooke et al, "Physics-enhanced machine learning for virtual fluorescence microscopy," ArXiv 2020.
Project page with data and source code: https://deepimaging.io/virtual-fluoroscence/
We have used microscopes to discover new phenomena for hundreds of years. Thanks to the digital image sensor and computer, much of this discovery work has now started to become automated. A variety of "deep" machine learning algorithms now automatically process digital microscope images to find, classify and interpret relevant phenomena, such as the indications of disease, or the presence of certain cells in an assay, or even a fully automated diagnosis
Despite their automation, microscopes themselves have still changed relatively little - they are, for the most part, still optimized for a human viewer to peer through to examine a sample in detail, which presents a number of challenges in the clinic. The diagnosis of infection by the malaria parasite offers a good example. Due to their small size (approximately 1 micron or less), the malaria parasite (P. Falciparum) must be viewed under a high-resolution objective lens (typically with oil immersion). Unfortunately, such high-resolution lenses can only see a very small area, containing just a few dozen cells. As the infection density of the malaria parasite is relatively low, one must scan through at least 100 unique fields-of-view to find enough examples to offer a sound diagnosis. This is true either if a human or an algorithm is viewing the images of each specimen - hundreds of images are still needed, which leads to a bottleneck in the diagnosis pipeline.
The Computational Optics Lab is currently solving problems like the one above by creating new microscopes, which are designed by deep learning algorithms, to ensure that their captured image data contains a maximum amount of information for the algorithm's specific task. This is a joint hardware-software optimization effort. In effect, we hope to turn the microscope into an "intelligent" agent, whose goal is to physically probe each specimen to allow the computer to learn as much as possible from it. Different optimizable hardware components that our lab has or currently is exploring inlcude programmable illumination, the optical pathway, and the detector and data management pipeline. Here are some of our currentprojects related to this "learned sensing" effort:
2. Learned sensing for joint optimization of different microscope components
Associated papers:
K. Kim et al, "Multi-element microscope optimization by a learned sensing network with composite physical layers," Optics Letters 2020.
Project page with data and source code: https://deepimaging.io/projects/me-microscope/

In this work, we investigate an approach to jointly optimize multiple microscope settings, together with a classification network, for improved performance of automated image analysis tasks. We explore the interplay between optimization of programmable illumination and pupil transmission, using experimentally imaged blood smears for automated malaria parasite detection, to show that multi-element “learned sensing” outperforms its single-element counterpart. While not necessarily ideal for human interpretation, the network’s resulting low-resolution microscope images (20X-comparable) offer a machine learning network sufficient contrast to match the classification performance of corresponding high-resolution imagery (100X-comparable), pointing a path towards accurate automation over large fields-of-view.
3. Learned sensing for optimized microscope illumination
Associated papers:
"Learned sensing: jointly optimized microscope hardware for accurate image classification," Biomed. Opt. Express (2019)
"Learned Integrated Sensing Pipeline: Reconfigurable Metasurface Transceivers as Trainable Physical Layer in an Artificial Neural Network", Advanced Science (2019)
Project page with data and source code: http://deepimaging.io/learned_sensing_dnn/
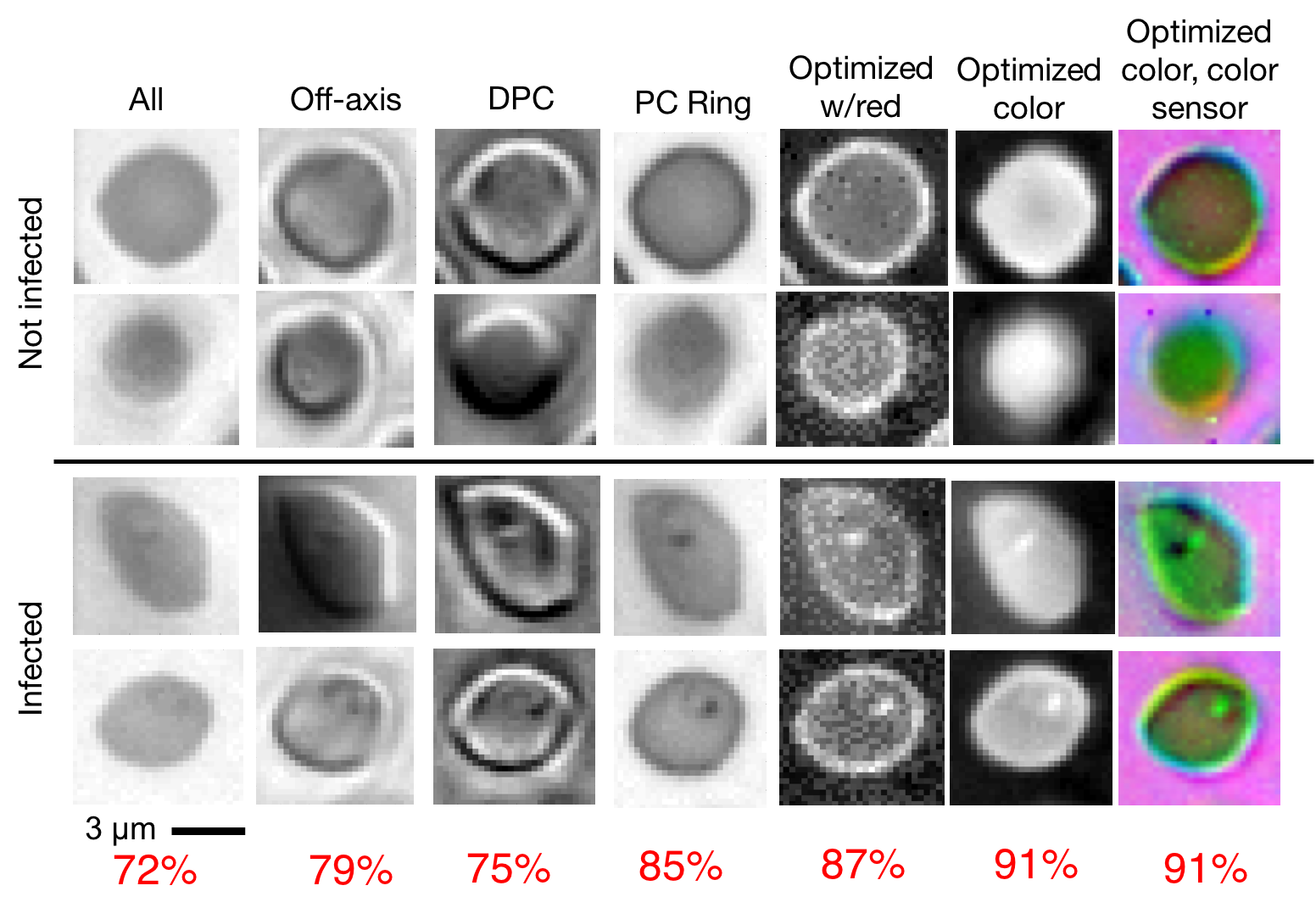
To significantly improve the speed and accuracy of disease diagnosis via light microscopy, we made two key modifications to the standard microscope: 1) we added a micro-LED unit that is optimized to illuminate each sample to highlight important features of interest (e.g., the malaria parasite within blood smears), and 2) we used a deep convolutional neural network to jointly optimize this illumination unit to automatically detect the presence of infection within the uniquely illuminated images.
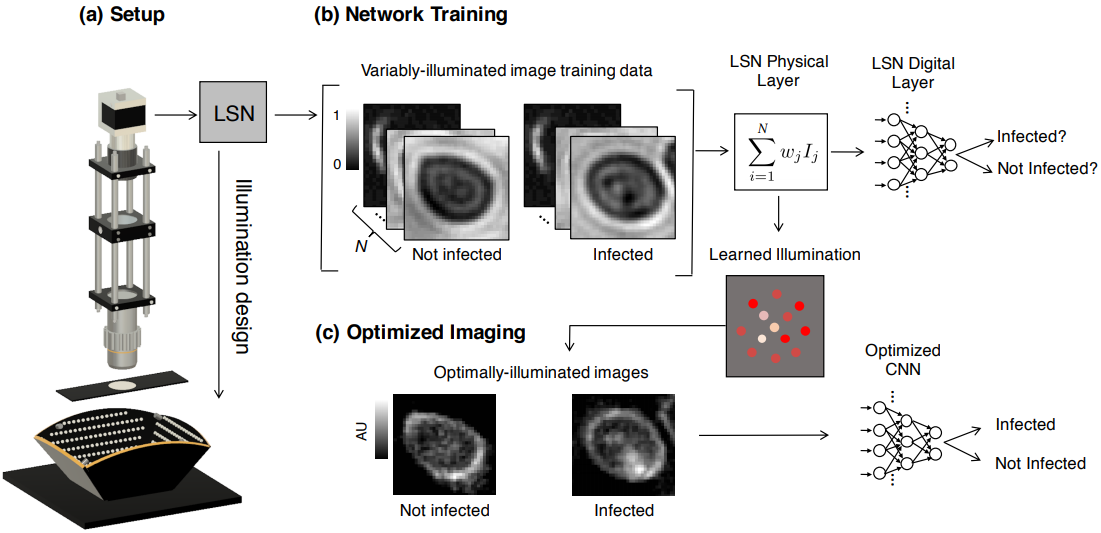
Working together, our two insights allow us to achieve classification accuracies in the 95 percentile range using large field-of-view, low-resolution microscope objective lenses that can see thousands of cells simultaneously (as opposed to just dozens of cells). This removes the need for mechanical scanning to obtain an accurate diagnosis, subsequently offering a dramatic speedup to the current diagnosis pipeline (i.e., from 10 minutes for manual inspection to just a few seconds for automatic inspection).
4. Adaptively learned illumination for optimal sample classification
Associated paper: "Towards an Intelligent Microscope: adaptively learned illumination for optimal sample classification," arXiv (2019)
Project page with data and source code: http://deepimaging.io/recurrent-illuminated-attention/

The Learned Sensing approach outlined above uses a convolutional neural network to establish optimized hardware settings. Here, we turned hardware optimization into a dynamic process, wherein we aim to teach the microscope how to interact with the specimen as it captures multiple images. To do so, we have turned to a reinforcement learning algorithm that treats the microscope as an agent, which can make dynamic decisions (how should I illuminate the sample next? How should I change the sample position? How should I filter the resulting scattered light?) on-the-fly during the image capture process.